A Connectome Computation System for discovery science of brain
Published in Science Bulletin, 2015
Recommended citation: Ting, Xu; Zhi, Yang; Lili, Jiang; Xiu-Xia, Xing; Xi-Nian, Zuo. (2015). "A Connectome Computation System for discovery science of brain." Science Bulletin. 60(1):86-95. https://github.com/zuoxinian/CCS/blob/master/manual/ccs.paper.pdf
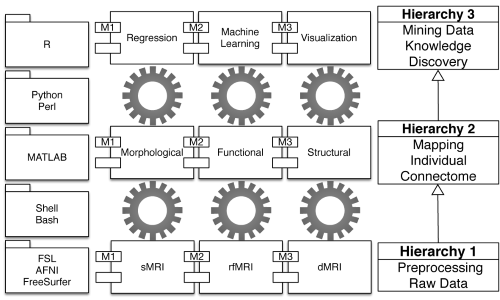
Much like genomics, brain connectomics has rapidly become a core component of most national brain projects around the world. Beyond the ambitious aims of these projects, a fundamental challenge is the need for an efficient, robust, reliable and easy-to-use pipeline to mine such large neuroscience datasets. Here, we introduce a computational pipeline—namely the Connectome Computation System (CCS)—for discovery science of human brain connectomes at the macroscale with multimodal magnetic resonance imaging technologies. The CCS is designed with a three-level hierarchical structure that includes data cleaning and preprocessing, individual connectome mapping and connectome mining, and knowledge discovery. Several functional modules are embedded into this hierarchy to implement quality control procedures, reliability analysis and connectome visualization. We demonstrate the utility of the CCS based upon a publicly available dataset, the NKI–Rockland Sample, to delineate the normative trajectories of well-known large-scale neural networks across the natural life span (6–85 years of age). The CCS has been made freely available to the public via GitHub to facilitate progress in discovery science in the field of human brain connectomics.
Recommended citation: Ting, Xu; Zhi, Yang; Lili, Jiang; Xiu-Xia, Xing; Xi-Nian, Zuo. (2015). “A Connectome Computation System for discovery science of brain.” Science Bulletin. 60(1):86-95.
